聚类kmeans算法在yolov3中的应用
yolov3 kmeans
yolov3在做boundingbox预测的时候,用到了anchor boxes.这个anchors的含义即最有可能的object的width,height.事先通过聚类得到.比如某一个像素单元,我想对这个像素单元预测出一个object,围绕这个像素单元,可以预测出无数种object的形状,并不是随便预测的,要参考anchor box的大小,即从已标注的数据中通过聚类统计到的最有可能的object的形状.
.cfg文件内的配置如下:
[yolo]
mask = 3,4,5
anchors = 10,14, 23,27, 37,58, 81,82, 135,169, 344,319
在用我们自己的数据做训练的时候,要先修改anchors,匹配我们自己的数据.anchors大小通过聚类得到.
通俗地说,聚类就是把挨得近的数据点划分到一起.
kmeans算法的思想很简单
- 随便指定k个cluster
- 把点划分到与之最近的一个cluster
- 上面得到的cluster肯定是不好的,因为一开始的cluster是乱选的嘛
- 更新每个cluster为当前cluster的点的均值.
这时候cluster肯定变准了,为什么呢?比如当前这个cluster里有3个点,2个点靠的很近,还有1个点离得稍微远点,那取均值的话,那相当于靠的很近的2个点有更多投票权,新算出来的cluster的中心会更加靠近这两个点.你要是非要抬杠:那万一一开始我随机指定的cluster中心点就特别准呢,重新取均值反而把中心点弄的不准了?事实上这是kmeans的一个缺陷:比较依赖初始的k个cluster的位置.选择不恰当的k值可能会导致糟糕的聚类结果。这也是为什么要进行特征检查来决定数据集的聚类数目了。 - 重新执行上述过程
- 把点划分到与之最近的一个cluster
- 更新每个cluster为当前cluster的点的均值
- 不断重复上述过程,直至cluster中心变化很小
yolov3要求的label文件格式
<object-class> <x_center> <y_center> <width> <height>
Where:
<object-class> - integer object number from 0 to (classes-1)
<x_center> <y_center> <width> <height> - float values relative to width and height of image, it can be equal from (0.0 to 1.0]
> for example: <x> = <absolute_x> / <image_width> or <height> = <absolute_height> / <image_height>
atention: <x_center> <y_center> - are center of rectangle (are not top-left corner)
举例:
1 0.716797 0.395833 0.216406 0.147222
所有的值都是比例.(中心点x,中心点y,目标宽,目标高)
kmeans实现
一般来说,计算样本点到质心的距离的时候直接算的是两点之间的距离,然后将样本点划归为与之距离最近的一个质心.
在yolov3中样本点的数据是有具体的业务上的含义的,我们其实最终目的是想知道最有可能的object对应的bounding box的形状是什么样子的. 所以这个距离的计算我们并不是直接算两点之间的距离,我们计算两个box的iou,即2个box的相似程度.d=1-iou(box1,box_cluster). 这样d越小,说明box1与box_cluster越类似.将box划归为box_cluster.
数据加载
f = open(args.filelist)
lines = [line.rstrip('\n') for line in f.readlines()]
annotation_dims = []
size = np.zeros((1,1,3))
for line in lines:
#line = line.replace('images','labels')
#line = line.replace('img1','labels')
line = line.replace('JPEGImages','labels')
line = line.replace('.jpg','.txt')
line = line.replace('.png','.txt')
print(line)
f2 = open(line)
for line in f2.readlines():
line = line.rstrip('\n')
w,h = line.split(' ')[3:]
#print(w,h)
annotation_dims.append(tuple(map(float,(w,h))))
annotation_dims = np.array(annotation_dims)
看着一大段,其实重点就一句
w,h = line.split(' ')[3:]
annotation_dims.append(tuple(map(float,(w,h))))
这里涉及到了python的语法,map用法https://www.runoob.com/python/python-func-map.html
这样就生成了一个N*2矩阵. N代表你的样本个数.
- 定义样本点到质心点的距离
计算样本x代表的box和k个质心box的IOU.(即比较box之间的形状相似程度).
这里涉及到一个IOU的概念:即交并集比例.交叉面积/总面积.
def IOU(x,centroids):
similarities = []
k = len(centroids)
for centroid in centroids:
c_w,c_h = centroid
w,h = x
if c_w>=w and c_h>=h: #box(c_w,c_h)完全包含box(w,h)
similarity = w*h/(c_w*c_h)
elif c_w>=w and c_h<=h: #box(c_w,c_h)宽而扁平
similarity = w*c_h/(w*h + (c_w-w)*c_h)
elif c_w<=w and c_h>=h:
similarity = c_w*h/(w*h + c_w*(c_h-h))
else: #means both w,h are bigger than c_w and c_h respectively
similarity = (c_w*c_h)/(w*h)
similarities.append(similarity) # will become (k,) shape
return np.array(similarities)
kmeans实现
def kmeans(X,centroids,eps,anchor_file):
N = X.shape[0]
iterations = 0
k,dim = centroids.shape
prev_assignments = np.ones(N)*(-1)
iter = 0
old_D = np.zeros((N,k)) #距离矩阵 N个点,每个点到k个质心 共计N*K个距离
while True:
D = []
iter+=1
for i in range(N):
d = 1 - IOU(X[i],centroids) #d是一个k维的
D.append(d)
D = np.array(D) # D.shape = (N,k)
print("iter {}: dists = {}".format(iter,np.sum(np.abs(old_D-D))))
#assign samples to centroids
assignments = np.argmin(D,axis=1) #返回每一行的最小值的下标.即当前样本应该归为k个质心中的哪一个质心.
if (assignments == prev_assignments).all() : #质心已经不再变化
print("Centroids = ",centroids)
write_anchors_to_file(centroids,X,anchor_file)
return
#calculate new centroids
centroid_sums=np.zeros((k,dim),np.float) #(k,2)
for i in range(N):
centroid_sums[assignments[i]]+=X[i] #将每一个样本划分到对应质心
for j in range(k):
centroids[j] = centroid_sums[j]/(np.sum(assignments==j)) #更新质心
prev_assignments = assignments.copy()
old_D = D.copy()
- 计算每个样本点到每一个cluster质心的距离 d = 1- IOU(X[i],centroids)表示样本点到每个cluster质心的距离.
- np.argmin(D,axis=1)得到每一个样本点离哪个cluster质心最近
argmin函数用法参考https://docs.scipy.org/doc/numpy/reference/generated/numpy.argmin.html - 计算每一个cluster中的样本点总和,取平均,更新cluster质心.
for i in range(N):
centroid_sums[assignments[i]]+=X[i] #将每一个样本划分到对应质心
for j in range(k):
centroids[j] = centroid_sums[j]/(np.sum(assignments==j)) #更新质心
- 不断重复上述过程,直到质心不再变化 聚类完成.
保存聚类得到的anchor box大小
def write_anchors_to_file(centroids,X,anchor_file):
f = open(anchor_file,'w')
anchors = centroids.copy()
print(anchors.shape)
for i in range(anchors.shape[0]):
anchors[i][0]*=width_in_cfg_file/32.
anchors[i][1]*=height_in_cfg_file/32.
widths = anchors[:,0]
sorted_indices = np.argsort(widths)
print('Anchors = ', anchors[sorted_indices])
for i in sorted_indices[:-1]:
f.write('%0.2f,%0.2f, '%(anchors[i,0],anchors[i,1]))
#there should not be comma after last anchor, that's why
f.write('%0.2f,%0.2f\n'%(anchors[sorted_indices[-1:],0],anchors[sorted_indices[-1:],1]))
f.write('%f\n'%(avg_IOU(X,centroids)))
print()
由于yolo要求的label文件中,填写的是相对于width,height的比例.所以得到的anchor box的大小要乘以模型输入图片的尺寸.
上述代码里
anchors[i][0]*=width_in_cfg_file/32.
anchors[i][1]*=height_in_cfg_file/32.
这里除以32是yolov2的算法要求. yolov3实际上不需要!!,注意你自己用的是yolov2还是v3,v3的话把/32去掉.参见以下链接https://github.com/pjreddie/darknet/issues/911
for Yolo v2: width=704 height=576 in cfg-file
./darknet detector calc_anchors data/hand.data -num_of_clusters 5 -width 22 -height 18 -show
for Yolo v3: width=704 height=576 in cfg-file
./darknet detector calc_anchors data/hand.data -num_of_clusters 9 -width 704 -height 576 -show
And you can use any images with any sizes.
完整代码见https://github.com/AlexeyAB/darknet/tree/master/scripts
用法:python3 gen_anchors.py -filelist ../build/darknet/x64/data/park_train.txt
/20190822***************/
完整代码 详细注释
'''
'''
Created on Feb 20, 2017
@author: jumabek
'''
from os import listdir
from os.path import isfile, join
import argparse
#import cv2
import numpy as np
import sys
import os
import shutil
import random
import math
width_in_cfg_file = 320.
height_in_cfg_file = 320.
def IOU(x,centroids):
similarities = []
k = len(centroids)
for centroid in centroids:
c_w,c_h = centroid
w,h = x
if c_w>=w and c_h>=h:
similarity = w*h/(c_w*c_h)
elif c_w>=w and c_h<=h:
similarity = w*c_h/(w*h + (c_w-w)*c_h)
elif c_w<=w and c_h>=h:
similarity = c_w*h/(w*h + c_w*(c_h-h))
else: #means both w,h are bigger than c_w and c_h respectively
similarity = (c_w*c_h)/(w*h)
similarities.append(similarity) # will become (k,) shape
return np.array(similarities)
def avg_IOU(X,centroids):
n,d = X.shape
sum = 0.
for i in range(X.shape[0]):
#note IOU() will return array which contains IoU for each centroid and X[i] // slightly ineffective, but I am too lazy
sum+= max(IOU(X[i],centroids))
return sum/n
def write_anchors_to_file(centroids,X,anchor_file):
f = open(anchor_file,'w')
anchors = centroids.copy()
print(anchors.shape)
for i in range(anchors.shape[0]):
anchors[i][0]*=width_in_cfg_file/32.
anchors[i][1]*=height_in_cfg_file/32.
widths = anchors[:,0]
sorted_indices = np.argsort(widths)
print('Anchors = ', anchors[sorted_indices])
for i in sorted_indices[:-1]:
f.write('%0.2f,%0.2f, '%(anchors[i,0],anchors[i,1]))
#there should not be comma after last anchor, that's why
f.write('%0.2f,%0.2f\n'%(anchors[sorted_indices[-1:],0],anchors[sorted_indices[-1:],1]))
f.write('%f\n'%(avg_IOU(X,centroids)))
print()
def kmeans(X,centroids,eps,anchor_file):
"""
X.shape = N * dim N代表全部样本数量,dim代表样本有dim个维度
centroids.shape = k * dim k代表聚类的cluster数,dim代表样本维度
"""
print("X.shape=",X.shape,"centroids.shape=",centroids.shape)
N = X.shape[0]
iterations = 0
k,dim = centroids.shape
prev_assignments = np.ones(N)*(-1)
iter = 0
old_D = np.zeros((N,k))
while True:
"""
D.shape = N * k N代表全部样本数量,k列分别为到k个质心的距离
1. 计算出D
2. 获取出当前样本应该归属哪个cluster
assignments = np.argmin(D,axis=1)
assignments.shape = N * 1 N代表N个样本,1列为当前归属哪个cluster
numpy里row=0,line=1,np.argmin(D,axis=1)即沿着列的方向,即每一行的最小值的下标
3. 将样本划分到相对应的cluster后,重新计算每个cluster的质心
centroid_sums.shape = k * dim k代表刻个cluster,dim列分别为该cluster的样本在该维度的均值
centroid_sums=np.zeros((k,dim),np.float)
for i in range(N):
centroid_sums[assignments[i]]+=X[i] # assignments[i]为cluster x 将每一个样本都归到其所属的cluster.
for j in range(k):
centroids[j] = centroid_sums[j]/(np.sum(assignments==j)) #np.sum(assignments==j)为cluster j中的样本总量
"""
D = []
iter+=1
for i in range(N):
d = 1 - IOU(X[i],centroids)
D.append(d)
D = np.array(D) # D.shape = (N,k)
print("iter {}: dists = {}".format(iter,np.sum(np.abs(old_D-D))))
assignments = np.argmin(D,axis=1)
#每个样本归属的cluster都不再变化了,就退出
if (assignments == prev_assignments).all() :
print("Centroids = ",centroids)
write_anchors_to_file(centroids,X,anchor_file)
return
#calculate new centroids
centroid_sums=np.zeros((k,dim),np.float)
for i in range(N):
centroid_sums[assignments[i]]+=X[i]
for j in range(k):
print("cluster{} has {} sample".format(j,np.sum(assignments==j)))
centroids[j] = centroid_sums[j]/(np.sum(assignments==j))
prev_assignments = assignments.copy()
old_D = D.copy()
def main(argv):
parser = argparse.ArgumentParser()
parser.add_argument('-filelist', default = '\\path\\to\\voc\\filelist\\train.txt',
help='path to filelist\n' )
parser.add_argument('-output_dir', default = 'generated_anchors/anchors', type = str,
help='Output anchor directory\n' )
parser.add_argument('-num_clusters', default = 0, type = int,
help='number of clusters\n' )
args = parser.parse_args()
if not os.path.exists(args.output_dir):
os.mkdir(args.output_dir)
f = open(args.filelist)
lines = [line.rstrip('\n') for line in f.readlines()]
#将label文件里的obj的w_ratio,h_ratio存储到annotation_dims
annotation_dims = []
for line in lines:
#line = line.replace('images','labels')
#line = line.replace('img1','labels')
line = line.replace('JPEGImages','labels')
line = line.replace('.jpg','.txt')
line = line.replace('.png','.txt')
print(line)
f2 = open(line)
for line in f2.readlines():
line = line.rstrip('\n')
w,h = line.split(' ')[3:]
#print(w,h)
annotation_dims.append(tuple(map(float,(w,h))))
annotation_dims = np.array(annotation_dims)
eps = 0.005
if args.num_clusters == 0:
for num_clusters in range(1,11): #we make 1 through 10 clusters
print(num_clusters)
anchor_file = join( args.output_dir,'anchors%d.txt'%(num_clusters))
indices = [ random.randrange(annotation_dims.shape[0]) for i in range(num_clusters)]
centroids = annotation_dims[indices]
kmeans(annotation_dims,centroids,eps,anchor_file)
print('centroids.shape', centroids.shape)
else:
anchor_file = join( args.output_dir,'anchors%d.txt'%(args.num_clusters))
##随机选取args.num_clusters个质心
indices = [ random.randrange(annotation_dims.shape[0]) for i in range(args.num_clusters)]
print("indices={}".format(indices))
centroids = annotation_dims[indices]
print("centroids=",centroids)
##
kmeans(annotation_dims,centroids,eps,anchor_file)
print('centroids.shape', centroids.shape)
if __name__=="__main__":
main(sys.argv)
如果训练图片的目标形状很少,比如就2,3种,那很可能
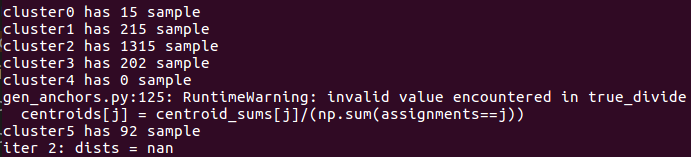
说明你的cluster过多了,某个cluster根本没有任何样本归属于他.那你可以通过命令行指定num_clusters.
python3 gen_anchors.py -filelist ./train.txt -num_clusters 3
最新文章
- 2016年Q2《网络安全创新500强》榜单解读
- IIS如何设置可以让.aspx后缀的文件直接下载
- 10g 11g配置Logical Standby
- script标签不带属性与带async、defer的区别
- 学会使用Constant常量或者Enum枚举
- Write operations are not allowed in read-only mode错误
- 1136. Parliament(二叉树)
- CentOS 6.5 IP 设置
- 将指定目录中的txt文件转化成excel文件
- SqlHelper工具类
- Tomcat初应用
- 屏幕旋转时调用PopupWindow update方法更新位置失效的问题及解决方案
- Hive 导入 parquet 格式数据
- SQL CTE递归
- 牛客网数据库SQL实战(此处只有答案,没有表内容)
- Mysql 索引问题-日期索引使用
- (转)Android学习路线总结,绝对干货
- 记一次使用mybatis进行like 模糊查询遇到的问题
- 关于Linux DNS部分处理
- SQL运算符