POJ 1080 Human Gene Functions -- 动态规划(最长公共子序列)
题目地址:http://poj.org/problem?id=1080
Description
their functions, because these can be used to diagnose human diseases and to design new drugs for them.
A human gene can be identified through a series of time-consuming biological experiments, often with the help of computer programs. Once a sequence of a gene is obtained, the next job is to determine its function.
One of the methods for biologists to use in determining the function of a new gene sequence that they have just identified is to search a database with the new gene as a query. The database to be searched stores many gene sequences and their functions – many
researchers have been submitting their genes and functions to the database and the database is freely accessible through the Internet.
A database search will return a list of gene sequences from the database that are similar to the query gene.
Biologists assume that sequence similarity often implies functional similarity. So, the function of the new gene might be one of the functions that the genes from the list have. To exactly determine which one is the right one another series of biological experiments
will be needed.
Your job is to make a program that compares two genes and determines their similarity as explained below. Your program may be used as a part of the database search if you can provide an efficient one.
Given two genes AGTGATG and GTTAG, how similar are they? One of the methods to measure the similarity
of two genes is called alignment. In an alignment, spaces are inserted, if necessary, in appropriate positions of
the genes to make them equally long and score the resulting genes according to a scoring matrix.
For example, one space is inserted into AGTGATG to result in AGTGAT-G, and three spaces are inserted into GTTAG to result in –GT--TAG. A space is denoted by a minus sign (-). The two genes are now of equal
length. These two strings are aligned:
AGTGAT-G
-GT--TAG
In this alignment, there are four matches, namely, G in the second position, T in the third, T in the sixth, and G in the eighth. Each pair of aligned characters is assigned a score according to the following scoring matrix.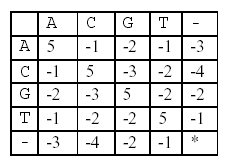
denotes that a space-space match is not allowed. The score of the alignment above is (-3)+5+5+(-2)+(-3)+5+(-3)+5=9.
Of course, many other alignments are possible. One is shown below (a different number of spaces are inserted into different positions):
AGTGATG
-GTTA-G
This alignment gives a score of (-3)+5+5+(-2)+5+(-1) +5=14. So, this one is better than the previous one. As a matter of fact, this one is optimal since no other alignment can have a higher score. So, it is said that the
similarity of the two genes is 14.
Input
The length of each gene sequence is at least one and does not exceed 100.
Output
Sample Input
2
7 AGTGATG
5 GTTAG
7 AGCTATT
9 AGCTTTAAA
Sample Output
14
21
#include <stdio.h>
int matrix[5][5] = {
{5, -1, -2, -1, -3},
{-1, 5, -3, -2, -4},
{-2, -3, 5, -2, -2},
{-1, -2, -2, 5, -1},
{-3, -4, -2, -1, 0},
};
char exchange[5] = {'A', 'C', 'G', 'T', ' '};
//通过矩阵求每一对字符的分值
int Value (char m, char n){
int R, C;
int i;
for (i=0; i<5; ++i){
if (exchange[i] == m)
R = i;
if (exchange[i] == n)
C = i;
}
return matrix[R][C];
}
int Max (int a, int b, int c){
int max = (a > b) ? a : b;
return (max > c) ? max : c;
}
int Similarity (char str1[], int length1, char str2[], int length2){
int dp[110][110];
int i, j;
dp[0][0] = 0;
for (i=1; i<=length1; ++i)
dp[i][0] = dp[i-1][0] + Value (str1[i], ' ');
for (i=1; i<=length2; ++i)
dp[0][i] = dp[0][i-1] + Value (' ', str2[i]);
///////////////////////////////////////////////////////////////////////////
//dp[i][j]表示第一个字串的长为i的子串与第二个字串的长为j的子串的相似度
for (i=1; i<=length1; ++i){
for (j=1; j<=length2; ++j){
///////////////////////////////////////////////////////////////////
//状态转移方程
if (str1[i] == str2[j]){
dp[i][j] = dp[i-1][j-1] + Value (str1[i], str2[j]);
}
else{
dp[i][j] = Max (dp[i-1][j] + Value (str1[i], ' '),
dp[i][j-1] + Value (' ', str2[j]),
dp[i-1][j-1] + Value (str1[i], str2[j]));
}
///////////////////////////////////////////////////////////////////
}
}
///////////////////////////////////////////////////////////////////////////
return dp[length1][length2];
}
int main(void){
char str1[110];
char str2[110];
int length1;
int length2;
int T;
int ans;
while (scanf ("%d", &T) != EOF){
while (T-- != 0){
scanf ("%d%s", &length1, str1 + 1);
scanf ("%d%s", &length2, str2 + 1);
if (length1 > length2)
ans = Similarity (str1, length1, str2, length2);
else
ans = Similarity (str2, length2, str1, length1);
printf ("%d\n", ans);
}
}
return 0;
}
HDOJ上相似的题目:http://acm.hdu.edu.cn/showproblem.php?pid=1513
最新文章
- Aop动态生成代理类时支持带参数构造函数
- mysql复习相关
- jQuery下拉框扩展和美化插件Chosen
- tomcat架构分析 (Session管理)
- Win7 64位 VS2015及MinGW环境编译矢量库agg-2.5和cairo-1.14.6
- Atitit.图片木马的原理与防范 attilax 总结
- Python 多线程 队列 示例
- Varnish 4.0
- Python小爬虫
- ASP.NET Core中使用GraphQL - 第二章 中间件
- 如何用EFCore Lazy Loading实现Entity Split
- java第一天 数据类型、变量的命名、类型的转换
- NPOI 操作excel之 将图片插入到指定位置;
- 内联外联CSS和JS
- ROW_NUMBER() OVER(PARTITION BY ORDER BY )RN 只选一行
- Tensorflow CNN入门
- Kafka网络模型分析
- Unity利用UI的Mask实现对精灵Sprite的遮挡
- asp.net和.net的区别
- composer gitlab 搭建私包
热门文章
- Linux 下 expect 脚本语言中交互处理常用命令
- Python Telnet弱口令爆破脚本及遇到的错误与问题
- js获取上传文件内容(未完待续)
- cocos2d-x Menu、MenuItem
- 并发与同步 (一) ThreadLocal与Synchronized 用哪一个好
- [Open Projects Series] ViewPagerTransforms
- android128 zhihuibeijing 科大讯飞 语音识别
- Mysql数据库导出压缩并保存到指定位置备份脚本
- C# 之 读取Word时发生 “拒绝访问” 及 “消息筛选器显示应用程序正在使用中” 异常的处理
- 获取mp4文件信息