Machine learning 第8周编程作业 K-means and PCA
2024-10-16 03:44:03
1.findClosestCentroids
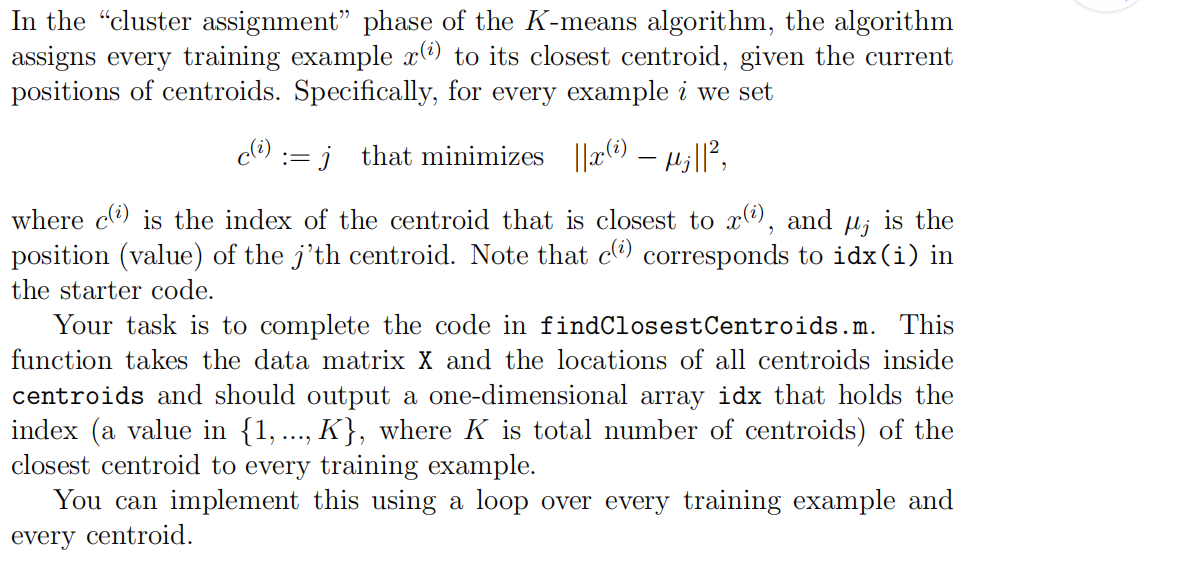
function idx = findClosestCentroids(X, centroids)
%FINDCLOSESTCENTROIDS computes the centroid memberships for every example
% idx = FINDCLOSESTCENTROIDS (X, centroids) returns the closest centroids
% in idx for a dataset X where each row is a single example. idx = m x 1
% vector of centroid assignments (i.e. each entry in range [1..K])
% % Set K
K = size(centroids, 1); % You need to return the following variables correctly.
idx = zeros(size(X,1), 1); % ====================== YOUR CODE HERE ======================
% Instructions: Go over every example, find its closest centroid, and store
% the index inside idx at the appropriate location.
% Concretely, idx(i) should contain the index of the centroid
% closest to example i. Hence, it should be a value in the
% range 1..K
%
% Note: You can use a for-loop over the examples to compute this.
% for i=1:size(X,1),
for j=1:K,
dis(j)=sum( (centroids(j,:)-X(i,:)).^2, 2 );
endfor
[t,idx(i)]=min(dis);
endfor % ============================================================= end
2.computerCentroids

function centroids = computeCentroids(X, idx, K)
%COMPUTECENTROIDS returns the new centroids by computing the means of the
%data points assigned to each centroid.
% centroids = COMPUTECENTROIDS(X, idx, K) returns the new centroids by
% computing the means of the data points assigned to each centroid. It is
% given a dataset X where each row is a single data point, a vector
% idx of centroid assignments (i.e. each entry in range [1..K]) for each
% example, and K, the number of centroids. You should return a matrix
% centroids, where each row of centroids is the mean of the data points
% assigned to it.
% % Useful variables
[m n] = size(X); % You need to return the following variables correctly.
centroids = zeros(K, n); % ====================== YOUR CODE HERE ======================
% Instructions: Go over every centroid and compute mean of all points that
% belong to it. Concretely, the row vector centroids(i, :)
% should contain the mean of the data points assigned to
% centroid i.
%
% Note: You can use a for-loop over the centroids to compute this.
% for i=1:K,
ALL=0;
cnt=sum(idx==i);
temp=find(idx==i);
for j=1:numel(temp),
ALL=ALL+X(temp(j),:);
endfor
centroids(i,:)=ALL/cnt;
endfor % ============================================================= end
3.pca
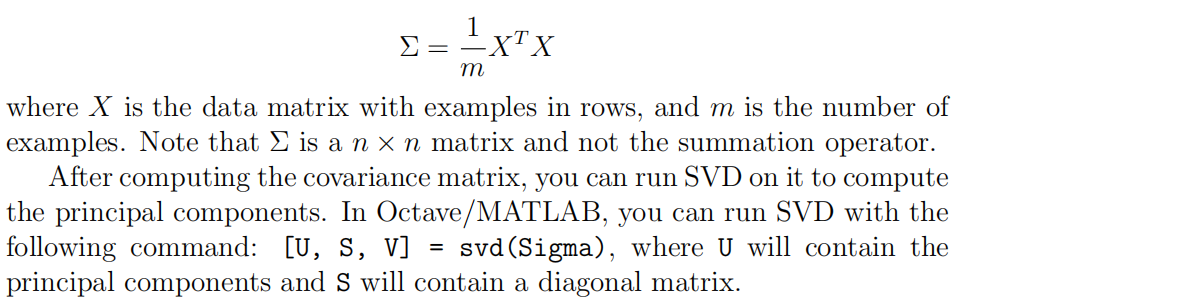
function [U, S] = pca(X)
%PCA Run principal component analysis on the dataset X
% [U, S, X] = pca(X) computes eigenvectors of the covariance matrix of X
% Returns the eigenvectors U, the eigenvalues (on diagonal) in S
% % Useful values
[m, n] = size(X); % You need to return the following variables correctly.
U = zeros(n);
S = zeros(n); % ====================== YOUR CODE HERE ======================
% Instructions: You should first compute the covariance matrix. Then, you
% should use the "svd" function to compute the eigenvectors
% and eigenvalues of the covariance matrix.
%
% Note: When computing the covariance matrix, remember to divide by m (the
% number of examples).
% Sigma=(X'*X)./m;
[U,S,V]=svd(Sigma); % ========================================================================= end
4.projectData
function Z = projectData(X, U, K)
%PROJECTDATA Computes the reduced data representation when projecting only
%on to the top k eigenvectors
% Z = projectData(X, U, K) computes the projection of
% the normalized inputs X into the reduced dimensional space spanned by
% the first K columns of U. It returns the projected examples in Z.
% % You need to return the following variables correctly.
Z = zeros(size(X, 1), K); % ====================== YOUR CODE HERE ======================
% Instructions: Compute the projection of the data using only the top K
% eigenvectors in U (first K columns).
% For the i-th example X(i,:), the projection on to the k-th
% eigenvector is given as follows:
% x = X(i, :)';
% projection_k = x' * U(:, k);
% U_reduce=U(:,1:K);
Z=X*U_reduce; % ============================================================= end
5.recoverData
function X_rec = recoverData(Z, U, K)
%RECOVERDATA Recovers an approximation of the original data when using the
%projected data
% X_rec = RECOVERDATA(Z, U, K) recovers an approximation the
% original data that has been reduced to K dimensions. It returns the
% approximate reconstruction in X_rec.
% % You need to return the following variables correctly.
X_rec = zeros(size(Z, 1), size(U, 1)); % ====================== YOUR CODE HERE ======================
% Instructions: Compute the approximation of the data by projecting back
% onto the original space using the top K eigenvectors in U.
%
% For the i-th example Z(i,:), the (approximate)
% recovered data for dimension j is given as follows:
% v = Z(i, :)';
% recovered_j = v' * U(j, 1:K)';
%
% Notice that U(j, 1:K) is a row vector.
%
X_rec=Z*U(:,1:K)'; % ============================================================= end
最新文章
- httpModules与Http模块
- Ajax 文件上传
- Java for LeetCode 214 Shortest Palindrome
- 苹果开发者账号注册&真机调试
- asp.net zip 压缩传输
- svn is already under version control问题解决
- JS面向对象思想(OOP)
- [Mugeda HTML5技术教程之17] 理解Mugeda访问统计结果
- Greenplum+Hadoop学习笔记-14-定义数据库对象之创建与管理模式
- parseSdkContent failed 解决方案
- HTML5原生拖拽/拖放⎡Drag & Drop⎦详解
- 图解HTTP,TCP,IP,MAC的关系
- linux和sqlserver 2017的安装
- LeetCode - 503. Next Greater Element II
- HDU1114(KB12-F DP)
- 自适应界面开发总结——WPF客户端开发
- 【转】Vim自动补全插件----YouCompleteMe安装与配置
- Java开发工程师(Web方向) - 01.Java Web开发入门 - 第3章.Tomcat
- AUC ROC PR曲线
- fis难用的地方
热门文章
- 在使用html5的video标签播放视频时为何只有声音却没有图像
- 2018.09.30 bzoj3551:Peaks加强版(dfs序+主席树+倍增+kruskal重构树)
- 2018.09.25 bzoj3572: [Hnoi2014]世界树(虚树+树形dp)
- spring boot web项目在IDEA下热部署解决办法(四步搞定)
- bootstrap-treeview的 简单使用
- AE IRasterCursor 获取栅格图层像素值
- (能被11整除的数的特征)The shortest problem --hdu
- PAT甲 1046. Shortest Distance (20) 2016-09-09 23:17 22人阅读 评论(0) 收藏
- HDU1241 Oil Deposits 2016-07-24 13:38 66人阅读 评论(0) 收藏
- DFS剪枝处理HDU1010