bcftools
beftools非常复杂,大概有20个命令,每个命令下面还有N多个参数
annotate .. edit VCF files, add or remove annotations call .. SNP/indel calling (former "view") cnv .. Copy Number Variation caller concat .. concatenate VCF/BCF files from the same set of samples consensus .. create consensus sequence by applying VCF variants convert .. convert VCF/BCF to other formats and back csq .. haplotype aware consequence caller filter .. filter VCF/BCF files using fixed thresholds gtcheck .. check sample concordance, detect sample swaps and contamination index .. index VCF/BCF isec .. intersections of VCF/BCF files merge .. merge VCF/BCF files files from non-overlapping sample sets mpileup .. multi-way pileup producing genotype likelihoods norm .. normalize indels plugin .. run user-defined plugin polysomy .. detect contaminations and whole-chromosome aberrations query .. transform VCF/BCF into user-defined formats reheader .. modify VCF/BCF header, change sample names roh .. identify runs of homo/auto-zygosity sort .. sort VCF/BCF files stats .. produce VCF/BCF stats (former vcfcheck) view .. subset, filter and convert VCF and BCF files
下面讲一下过滤参数
1、bcftools filter(以-i参数为例)
- -i, --include EXPRESSION :
- include only sites for which EXPRESSION is true. For valid expressions see EXPRESSIONS.(根据正则保留)
- 其中包括:
- 1.1、numerical constants, string constants, file names (this is currently supported only to filter by the ID column)
1, 1.0, 1e-4 "String" @file_name
1.2、算术运算
+,*,-,/
1.3、comparison operators
== (same as =), >, >=, <=, <, !=
1.4、regex操作符“~”和它的否定“!~”。表达式区分大小写,除非添加“/i”。
INFO/HAYSTACK ~ "needle" INFO/HAYSTACK ~ "NEEDless/i"
1.5、圆括号
(, )
1.6、逻辑运算符。参见下面的示例和有关“&&”与“&”以及“||”与“|”之间区别的过滤教程。
&&, &, ||, |
1.7、信息标签,格式标签,列名
INFO/DP or DP FORMAT/DV, FMT/DV, or DV FILTER, QUAL, ID, CHROM, POS, REF, ALT[0]
1.8、1 (or 0) to test the presence (or absence) of a flag
FlagA=1 && FlagB=0
1.9、"." to test missing values
DP=".", DP!=".", ALT="."
2.0、missing genotypes can be matched regardless of phase and ploidy (".|.", "./.", ".") using these expressions
GT~"\.", GT!~"\."
2.1、sample genotype: reference (haploid or diploid), alternate (hom or het, haploid or diploid), missing genotype, homozygous, heterozygous, haploid, ref-ref hom, alt-alt hom, ref-alt het, alt-alt het, haploid ref, haploid alt (case-insensitive)
GT="ref" GT="alt" GT="mis" GT="hom" GT="het" GT="hap" GT="RR" GT="AA" GT="RA" or GT="AR" GT="Aa" or GT="aA" GT="R" GT="A"
2.2、TYPE for variant type in REF,ALT columns (indel,snp,mnp,ref,bnd,other,overlap). Use the regex operator "\~" to require at least one allele of the given type or the equal sign "=" to require that all alleles are of the given type. Compare
TYPE="snp" TYPE~"snp" TYPE!="snp" TYPE!~"snp"
2.3、array subscripts (0-based), "*" for any element, "-" to indicate a range. Note that for querying FORMAT vectors, the colon ":" can be used to select a sample and an element of the vector, as shown in the examples below
INFO/AF[0] > 0.3 .. first AF value bigger than 0.3 FORMAT/AD[0:0] > 30 .. first AD value of the first sample bigger than 30 FORMAT/AD[0:1] .. first sample, second AD value FORMAT/AD[1:0] .. second sample, first AD value DP4[*] == 0 .. any DP4 value FORMAT/DP[0] > 30 .. DP of the first sample bigger than 30 FORMAT/DP[1-3] > 10 .. samples 2-4 FORMAT/DP[1-] < 7 .. all samples but the first FORMAT/DP[0,2-4] > 20 .. samples 1, 3-5 FORMAT/AD[0:1] .. first sample, second AD field FORMAT/AD[0:*], AD[0:] or AD[0] .. first sample, any AD field FORMAT/AD[*:1] or AD[:1] .. any sample, second AD field (DP4[0]+DP4[1])/(DP4[2]+DP4[3]) > 0.3 CSQ[*] ~ "missense_variant.*deleterious"
2.4、with many samples it can be more practical to provide a file with sample names, one sample name per line
GT[@samples.txt]="het" & binom(AD)<0.01
2.5、function on FORMAT tags (over samples) and INFO tags (over vector fields): maximum; minimum; arithmetic mean (AVG is synonymous with MEAN); median; standard deviation; sum; string length; absolute value; number of elements (matching columns for FORMAT tags or number of fields for INFO tags).
MAX, MIN, AVG, MEAN, MEDIAN, STDEV, SUM, STRLEN, ABS, COUNT
2.6、two-tailed binomial test. Note that for N=0 the test evaluates to a missing value and when FORMAT/GT is used to determine the vector indices, it evaluates to 1 for homozygous genotypes.
binom(FMT/AD) .. GT can be used to determine the correct index binom(AD[0],AD[1]) .. or the fields can be given explicitly phred(binom()) .. the same as binom but phred-scaled
2.7、variables calculated on the fly if not present: number of alternate alleles; number of samples; count of alternate alleles; minor allele count (similar to AC but is always smaller than 0.5); frequency of alternate alleles (AF=AC/AN); frequency of minor alleles (MAF=MAC/AN); number of alleles in called genotypes; number of samples with missing genotype; fraction of samples with missing genotype; indel length (deletions negative, insertions positive)
N_ALT, N_SAMPLES, AC, MAC, AF, MAF, AN, N_MISSING, F_MISSING, ILEN
2.8、the number (N_PASS) or fraction (F_PASS) of samples which pass the expression
N_PASS(GQ>90 & GT!="mis") > 90 F_PASS(GQ>90 & GT!="mis") > 0.9
2.9、custom perl filtering. Note that this command is not compiled in by default, see the section Optional Compilation with Perl in the INSTALL file for help and misc/demo-flt.pl for a working example. The demo defined the perl subroutine "severity" which can be invoked from the command line as follows:
perl:path/to/script.pl; perl.severity(INFO/CSQ) > 3
注意事项:
字符串比较和正则表达式不区分大小写;变量和函数名不区分大小写,但是flag区分大小写。例如,"qual"可以代替"qual", "strlen()"可以代替"strlen()",但不是“dp”而是“DP”。当查询多个值时,将测试所有元素并对结果使用OR逻辑。例如,查询“TAG=1,2,3,4”时,计算如下:
-i 'TAG[*]=1' .. true, the record will be printed -i 'TAG[*]!=1' .. true -e 'TAG[*]=1' .. false, the record will be discarded -e 'TAG[*]!=1' .. false -i 'TAG[0]=1' .. true -i 'TAG[0]!=1' .. false -e 'TAG[0]=1' .. false -e 'TAG[0]!=1' .. true
举例:
MIN(DV)>5 MIN(DV/DP)>0.3 MIN(DP)>10 & MIN(DV)>3 FMT/DP>10 & FMT/GQ>10 .. both conditions must be satisfied within one sample FMT/DP>10 && FMT/GQ>10 .. the conditions can be satisfied in different samples QUAL>10 | FMT/GQ>10 .. true for sites with QUAL>10 or a sample with GQ>10, but selects only samples with GQ>10 QUAL>10 || FMT/GQ>10 .. true for sites with QUAL>10 or a sample with GQ>10, plus selects all samples at such sites TYPE="snp" && QUAL>=10 && (DP4[2]+DP4[3] > 2) COUNT(GT="hom")=0 .. no homozygous genotypes at the site AVG(GQ)>50 .. average (arithmetic mean) of genotype qualities bigger than 50 ID=@file .. selects lines with ID present in the file ID!=@~/file .. skip lines with ID present in the ~/file MAF[0]<0.05 .. select rare variants at 5% cutoff POS>=100 .. restrict your range query, e.g. 20:100-200 to strictly sites with POS in that range.
shell 扩展:
注意表达式必须经常引用,因为在shell中有些字符有特殊的含义。一个用单引号括起来的表达式的例子,它导致整个表达式按照预期传递给程序:
bcftools view -i '%ID!="." & MAF[0]<0.01'
------------------------过滤Filtering-------------
1、按照固定列(fixed columns)过滤
固定列,例如“QUAL, FILTER, INFO”可以直接过滤。例如:
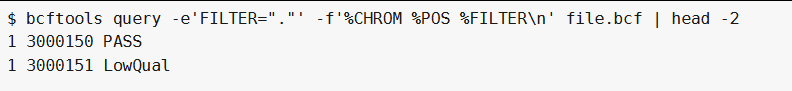
bcftools query -e'FILTER="."' -f'%CHROM %POS %FILTER\n' file.bcf #过滤掉FILTER字段中为.的行
bcftools query -i'QUAL>20 && DP>10' -f'%CHROM %POS %QUAL %DP\n' file.bcf | head -2 #只保留质量值大于20,且覆盖深度高于10的位点
2、FORMAT columns
在过滤FORMAT字段的时候,OR 逻辑用于所有samples。When filtering FORMAT tags, the OR logic is applied with multiple samples,而不是单个sample.例如,如果我们想删除任何样本中带有未知基因型的位点,表达式-i 'GT!="会不起作用,必须用相反的逻辑 -e 'GT ="."' :
bcftools query -i 'GT!="."' #不行 bcftools query -e 'GT ="."' #相反逻辑才可行

3、FORMAT列与 布尔值(&& vs & and || vs |)
我们希望一个sample或多个samples具有足够大的覆盖率(DP>10)和基因型质量(GQ>20)的snp位点:
bcftools query -i'FMT/DP>10 & FMT/GQ>20' -f'%POS[\t%SAMPLE:DP=%DP GQ=%GQ]\n' file.bcf ##-i 'FMT/DP>10和FMT/GQ>20'在同一个sample中选择满足条件的位点:

另一方面,如果我们需要在同一sample中两个条件都满足但不一定相同样品,我们使用&&操作符而不是&:
bcftools query -i'FMT/DP>10 && FMT/GQ>20' -f'%POS[\t%SAMPLE:DP=%DP GQ=%GQ]\n' file.bcf
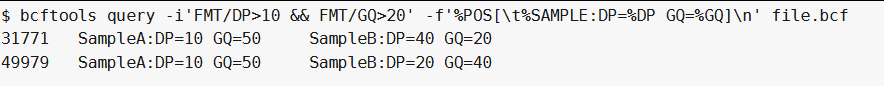
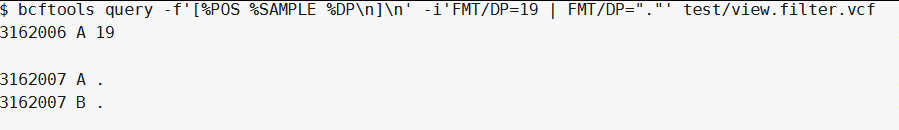
|操作符可以只选择匹配的样本:
bcftools query -f'[%POS %SAMPLE %DP\n]\n' -i 'FMT/DP=19 | FMT/DP="."' test/view.filter.vcf
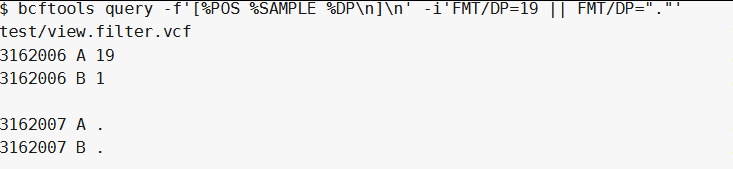
whole samples record when || is used(就是有一个样本符合该位点,那么该位点所有的样本记录都会被显示出来):
bcftools query -f '[%POS %SAMPLE %DP\n]\n' -i 'FMT/DP=19 || FMT/DP="."' test/view.filter.vcf
过滤:
bcftools filter -i 'SVLEN<100000 | SVLEN< -50 & DV>10' -Oz --threads 8 -o B1952.filter.clean.vcf.gz B1952.ngmlr_sniffle.vcf #例如过滤-50<绝对SVLEN<10000
重要网址:
http://samtools.github.io/bcftools/howtos/filtering.html
最新文章
- Command /usr/bin/codesign failed with exit code 1
- 【C解毒】滥用变量
- SEP图示
- 腾讯优测干货精选| 安卓开发新技能Get -常用必备小工具汇总
- Java开发之单例设计模式
- 【HDOJ】2150 Pipe
- 【数据库】SQL优化方法汇总
- fsck害了我很久了,必须关掉,因为他每次打卡都要推迟数十分钟。
- Python中库或者模组的解释
- Caused by: java.lang.ClassNotFoundException: org.springframework.orm.hibernate4.HibernateTemplate
- 《BUG创造队》第二次团队作业:团队项目选题报告
- dll 已注册 检索 COM 类工厂中 CLSID 为 {XXXX-XXXX-XXX-XXXXX-XXX} 的组件时失败,原因是出现以下错误: 80040154。
- [js] - 前端FileReader使用,适用于文件上传预览.(并未传入后端)
- mac电脑读写NTFS格式的移动硬盘命令
- 导入maven框架报错
- HBase写入性能改造(续)--MemStore、flush、compact参数调优及压缩卡的使用【转】
- shell脚本传递带有空格的参数的解决方法
- BZOJ 4826: [Hnoi2017]影魔 单调栈 主席树
- vim/vi 命令详解
- u3d性能优化
